Laboratory of Systembryogenesis
Unraveling the regulatory principles of embryogenesis through systems-level
analyses using in situ, single-cell approaches
analyses using in situ, single-cell approaches
Our team is dedicated to uncovering the intricacies of embryogenesis - the remarkable transformation of a single cell into a complete organism – through the lens of the C. elegans model. We adopt a cell-centric perspective on development, treating individual cells as functional and regulatory units, and employing systems biology strategies to decode the underlying regulatory mechanisms. Instead of focusing on specific genes, pathways, or processes, our research is driven by two core inquiries: How does genome information specify cellular states? And how do cellular states influence developmental functions? To tackle these questions, we utilize a multidisciplinary approach that integrates genetics, live-embryo imaging, functional genomics, phenomics, and systems biology. Our long-term goal is to understand the regulation of embryogenesis at the systems level.

The Making of an Embryo
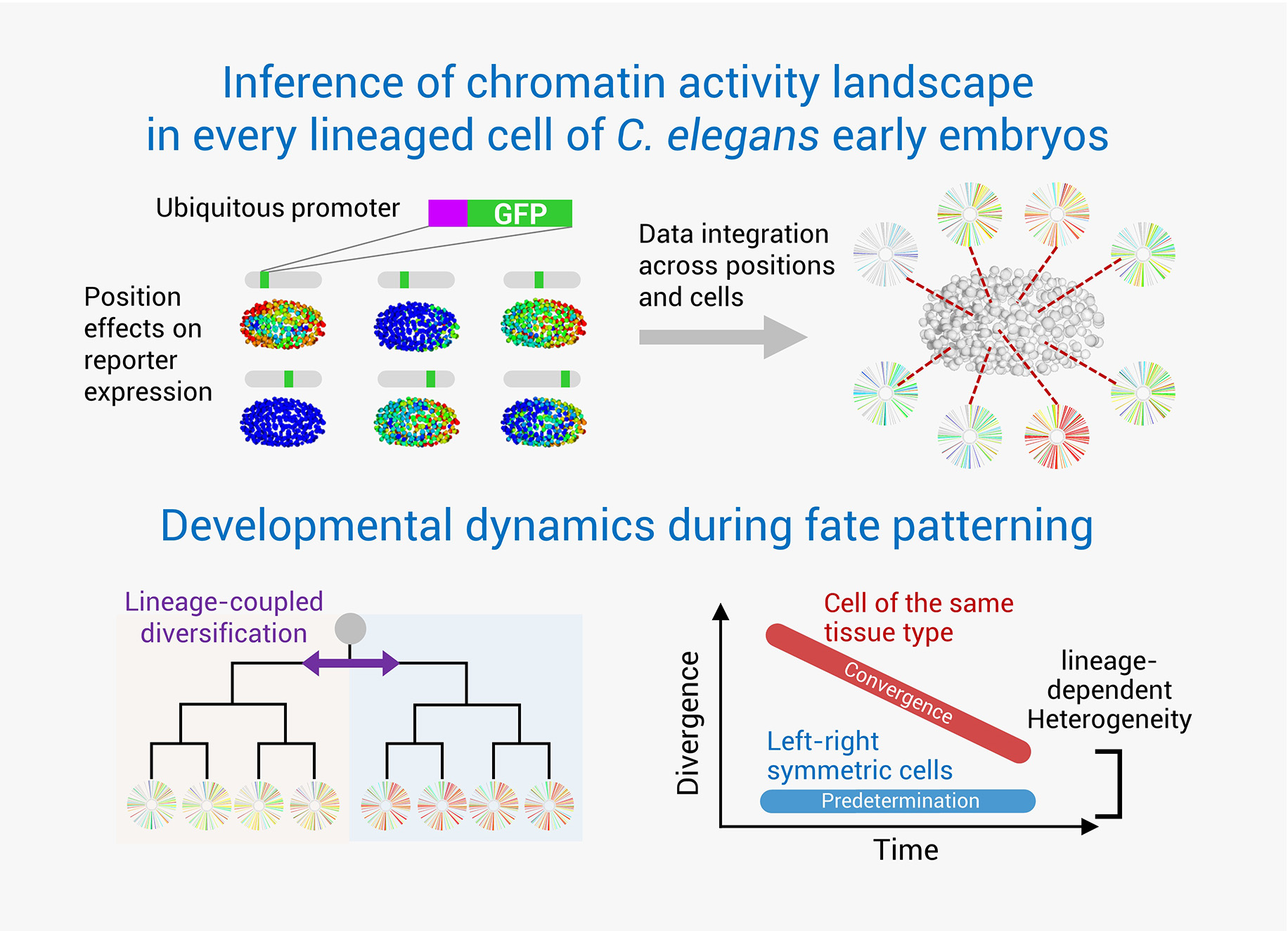
C. elegans has achieved a landmark status as the first multicellular organism to have its entire cell atlas mapped. This atlas encompasses the developmental history, anatomy, and ultimate fate of all 959 somatic cells that make up the organism. With advanced genetic tools, C. elegans stands at the forefront of research into development regulation, facilitating in-depth studies at both the single-cell and whole-organism scales. Despite the comprehensive resolution of the developmental cell atlas, a systematic exploration of the molecular dynamics in developing cells remains to be accomplished. The existing gaps include a comprehensive knowledge of the chromatin states, gene transcription profiles, protein dynamics, molecular interactions, and regulatory networks within each cell during development. Gaining insights into these processes is crucial for understanding normal development, as well as modeling and engineering cell fates and functions in cell therapy.
To bridge this knowledge gap, our lab employs a high-throughput, imaging-based approach for conducting single-cell functional analysis of embryogenesis. This technique utilizes long-term 3D time-lapse imaging to record embryonic development, coupled with systematic cell lineage tracing and quantitative analyses. Such an approach enables accurate quantification of each cell’s molecular activities and developmental dynamics, including aspects such as proliferation, gene expression, differentiated state, and spatial positioning. By integrating this powerful technique with system-wide genetic perturbations, we aim to unravel the intricate molecular mechanisms governing embryogenesis.
Ongoing projects include:
Novel single-cell methods to trace key regulatory events during embryogenesis
Spatiotemporal patterning codes of developmental fates
Regulation of developmental transition from multipotency to unipotency
Spatiotemporal patterning codes of developmental fates
Regulation of developmental transition from multipotency to unipotency
The Making of a Robust Embryo
Another intriguing aspect of embryogenesis is its remarkable robustness, which ensures individual embryos develop into morphologically and functionally consistent entities despite significant molecular, cellular, and environmental variation. Studies have unveiled critical molecular mechanisms that enhance the robustness of biological processes, including parallel pathways, stochastic expression variability, induced genetic compensations, and gene network topology. Although these molecular mechanisms shed light on how biological systems manage molecular noises, they fall short of directly explaining how developing systems tackle noise at the cellular level. Questions arise about how embryos deal with developmental noises at the cellular level, whether embryonic cells possess induced behavioral and functional plasticity as a means to buffer noise, and the role of cell-cell communication in achieving robust development.
C. elegans serves as a powerful model system to explore these questions, owing to its highly reproducible developmental processes. Its embryogenesis is characterized by an invariant cell lineage that produces an identical set of cells in the exact same manner across all individuals, allowing for the identification and comparison of equivalent cells and their phenotypes. Moreover, the predictability of the developmental behaviors of individual cells lends significant biological relevance to the study of cellular states and behavior noise. To gain a comprehensive understanding of developmental robustness across different scales, we employ C. elegans as the model to investigate how cellular-level mechanisms contribute to the robustness of multicellular developing systems.
Ongoing projects include:
Contributions of cellular compensation to developmental robustness
Genetic determinants of cellular plasticity
Genetic determinants of cellular plasticity