WormBase ID: WBGene00001311 WormBase description: Is predicted to have DNA-binding transcription factor activity, RNA polymerase II-specific and RNA polymerase II proximal promoter sequence-specific DNA binding activity. Is involved in endodermal cell fate specification and mesodermal cell fate specification. Localizes to nucleus. Is expressed in E; E lineage cell; Ea; Ep; and intestine. |
Strain name: SYS436 KI background: JIM113 Strain genotype: ujIs113 (pie-1p::mCherry::H2B; nhr-2p::mCherry::his-24::let858 3'UTR; unc-119(+)) II; end-3 (devKi109 [mNeonGreen::end-3]) V |
Expression pattern: Lineage-specific, Intestine-specific, Transient-expression |
Lineage expression pattern 350 stage 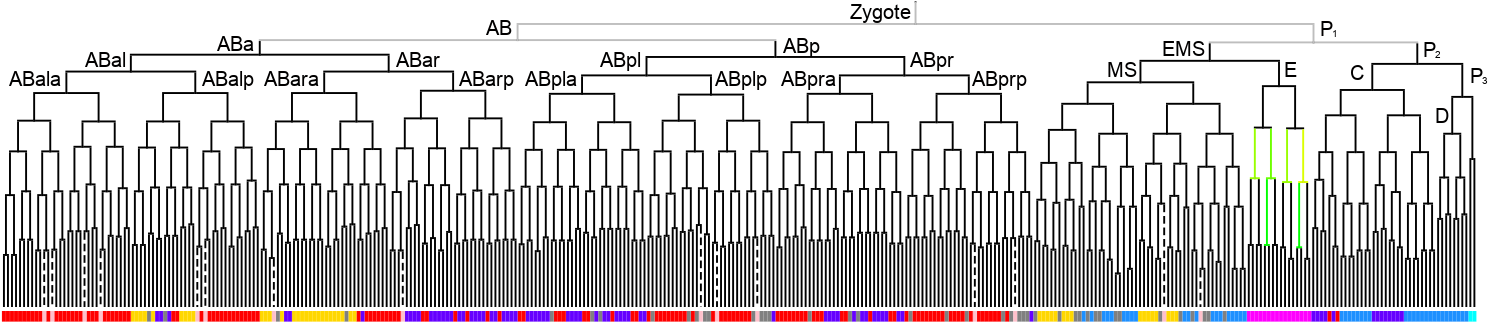 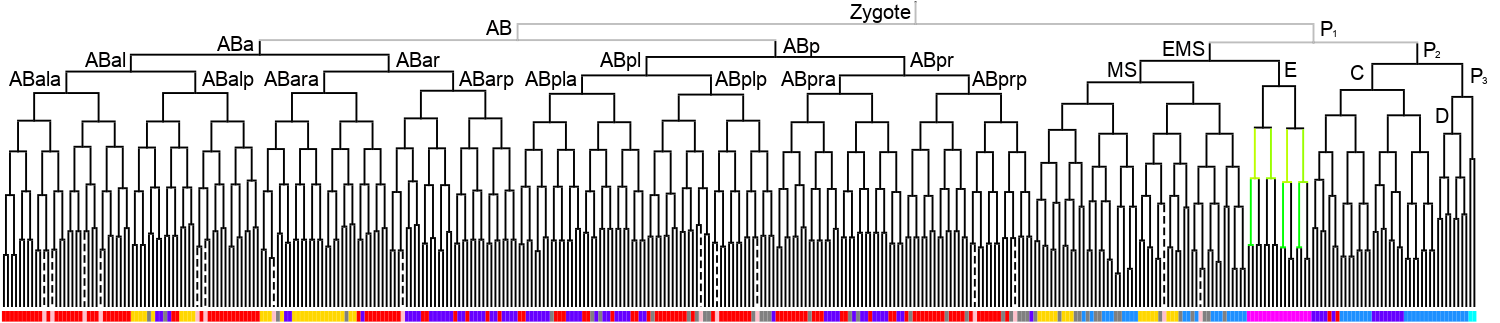 |
600 stage 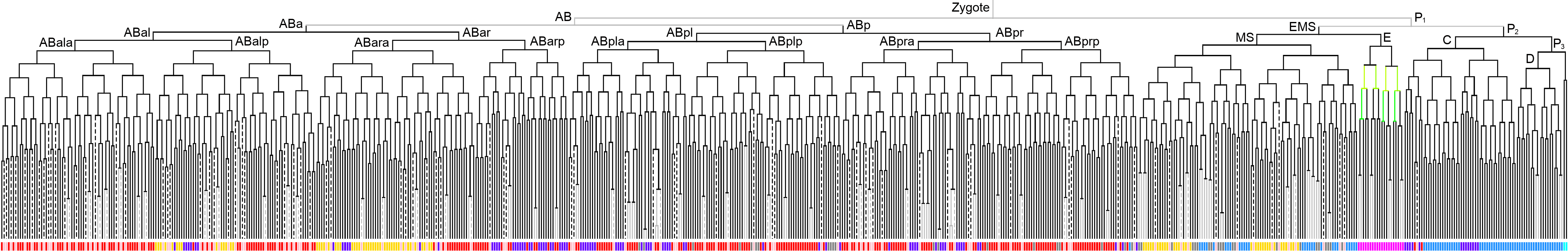 |
3D expression pattern  4-cells  8-cells  15-cells  28-cells  ~50-cells 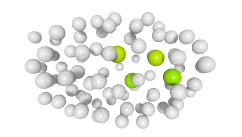 ~100-cells 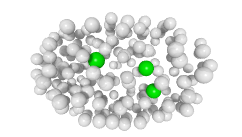 ~200-cells  ~350-cells  ~500-cells  ~600-cells  600-cells  bean-stage |
Inferred TF relationships Based on expression similarity (strong, medium, weak): END-1, ELT-7, MED-2, ELT-2, DVE-1, NHR-102, TBX-9, CEH-13, HMBX-1, TBX-11 Based on ChIP-seq (directly from ModERN database, targets from L1 stage ChIP-seq data are underlined) Targeting: no-data Targeted by: NA |
|
Targeting: no-data Targeted by: NA |