WormBase ID: WBGene00013583 WormBase description: Is an ortholog of human HOXB8 (homeobox B8). Is predicted to have DNA-binding transcription factor activity and RNA polymerase II proximal promoter sequence-specific DNA binding activity. Is involved in mesodermal cell fate specification; muscle cell fate specification; and pharyngeal muscle development. Localizes to nucleus. Is expressed in several structures, including MSaa; MSap; MSp; MSpa; and MSpp. Human ortholog(s) of this gene are implicated in amelogenesis imperfecta type 4; orofacial cleft 15; and split hand-foot malformation 1 with sensorineural hearing loss. |
Strain name: SYS397 KI background: JIM113 Strain genotype: ujIs113 (pie-1p::mCherry::H2B; nhr-2p::mCherry::his-24::let858 3'UTR; unc-119(+)) II; ceh-51 (devKi72 [mNeonGreen::ceh-51]) V |
Expression pattern: Lineage-specific, Transient-expression |
Lineage expression pattern 350 stage     |
600 stage 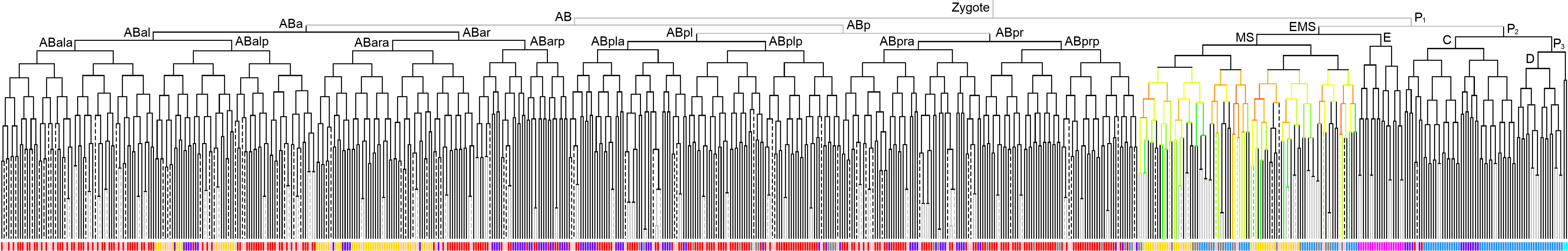 |
3D expression pattern  4-cells  8-cells  15-cells  28-cells  ~50-cells  ~100-cells  ~200-cells  ~350-cells  ~500-cells  ~600-cells  600-cells  bean-stage |
Inferred TF relationships Based on expression similarity (strong, medium, weak): LET-381, ODD-2, DMD-4, MED-2, TBX-7, HMBX-1, ALR-1, CEH-34, PHA-4, IRX-1, REF-2, M03D4.4 Based on ChIP-seq (directly from ModERN database, targets from L1 stage ChIP-seq data are underlined) Targeting: no-data Targeted by: NA |
|
Targeting: no-data Targeted by: PAG-3 |