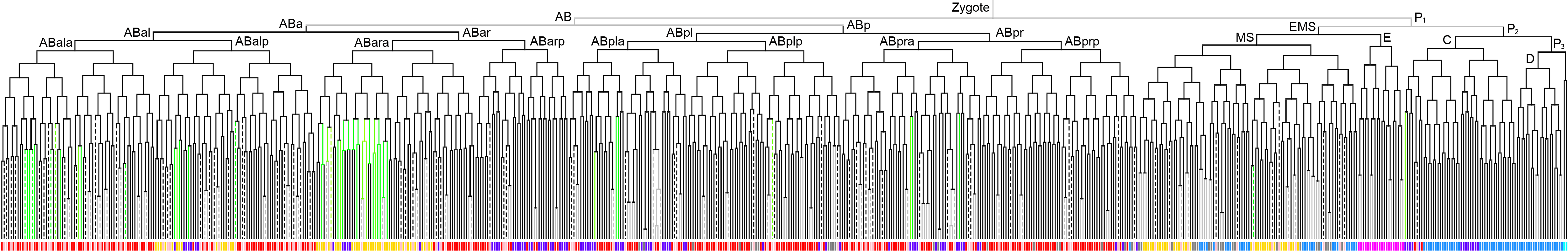
|
KI information: mNeonGreen knockin at N-terminus
sgRNA1: TCTGTAGGGATTTCGCTCAT
sgRNA2: ATGTTCAAGGGCATAATTTA
5'HR: GGATCCATGTAGCTGTGAAACACAATCCAAGAAGAGCAACCACGTCGGCATGACGTCAGTTCTCATTTTATTCTGTGGTTTTCCACGTCTCGATGGATCATCGTCGGTTCTTCCGAGTAGCCGTTCACCAAATGGGTGGCCAAGGGCAATCCACATTTTTTCGATGAGTTGTTGAAGACCTGAAATTGATTTTTAAATTGAAAAAAAAATTTCGGAAAATTTTGGAAATTAATACAAGGAGAGAACTAAATTAAAATTAGTTTTTTAAACCCCATTTTTAGTACAGAAAAAACATTTAATAAACTCACCCGTAGTTGTTCTGTAAAATGGATCACAGCACATTTGCGTGAGGCTGGCCACAATAATCGACATATCTGATCCTTCGTCTTCTGCAAAATATCCGGTGTTATAACCATTCAAAAGTACCATCAATTTGTTAGAGATAAACTCAAGATTAATGCATTTTAGTAGAAATCAGTAAGGGAAACTGTAAATCGTAGCCGAGTAGATCGAACTTCTTTCATAATTTTTTTACGATTGACCGGAGGTTTTTTCCATTTCTTGGTATTTTCTTTTCAATTCCAAAAAGTCATTTACATAACTACTTACATACAAATATGTGGGGTGCCCATACACGTATTCAGTCTTACTCTCTTTTCATATAATAATGTAGTTTTTCTATTTGTCTTGTCTCCTCCTATTAGCCTTGCACCCCCTTTACATTCTGACCAGTCGGAAATTCGTCTCACTGCCTCTACCCTCTAATAGTCTTGCACACTCTATTAGCCATGTCTCTCTGCGTGTGTGCCAATTTGCTGTCTCCATTAGTCTTGCCCCTTCGTTCGCTTTTCTCACATGTTTTATTTCCAATAACTTGCTGACTAGTTAAGACTTTGCTTGTTATATACTTATTTTTCAGAGAAAACCAGGTACC
3'HR: CTGCAGATGAGTGAAATCCCTACAGAAGTGCCTGTAAAAACTCGAAAAAGAACGTATGACTTGAAGTTCAAACTGCATGCTATAAACTATGCCCTTGAACATAAGAATATATCGAAAGCAGCAAAGGATTTGAATGTGAATAGGCAAAACATTCAACAGTGGATCGCCCAGAAAGCTGAAATCGAGAAGCAGTTGTAAGTTTTGCCACTTCGGACATATTTTTGTGTTCAAAAAAATAGTCCAGGGCTAGGACTTTCATGTTAGGCATGCGCGGTGTCAGGGTCCTGCTTGAAACCCGCCTATGTGAATTTACAAATTTCTAATTTTCTCATTTCTCTCAATTTGATGAAAATCAAAATAGAAATTAAAAAAAAAAAGATTGATTATGTTTTAGCACAACGTAAGGCGTAACCGAGAGGCAAGCGGAGGTCGCCTTAAAGGCAGGGTTTAACGCCTACATGCAAGCCCTATCAATTGAGAACCTTTTATGGACCTTTTATTCCAAATACTATGATTTTTTCTAGGCTTATAATTAATCCTATCAAAAGAAGTCATTTCAACTATTCCTATATTTTGAAATACGGGCATAACATAAATTCTCTGTTCGATTTCAAATTTTCTTTTTGAAAAATTTTCGACTTGCTGCCTTTTGGCAAATAACACTTTCAGAGAAGACAACGGCTCAACTGCAACAAAGCGACTTTCGGGAGGTGGAAGACCTTTAAAGTATGACGACGTTGATAAGGAATTGATTAAATGGGTTCACGATCAGCGTAAGGAGAAACAACGAGTCACCAGAAGAATCATCGGTGAAACGGCAAAGAAAATCAGTCAGAACTCGGATTTCAAGGTAGGAGAGCCGTTACTTTAATGCAAACGAGTATATGAAAAACATAAAAAATAACTATTCGACGCTACTCCACCTTTATCATTCAGGCATCCCGCGGATGGTTGGACACTAGT
KI verification primers: Y48G1C.6-NKIF: 5'-ACCAAACCTACTCAAATGTATATATTCG-3' Y48G1C.6-NKIR: 5'-TCCGTTCTTGCTCGTCGAGTA-3' Download
|
Inferred TF relationships
Based on expression similarity (strong, medium, weak): AHA-1, CEH-2, PHA-2, PAX-1, CEH-5, DMD-5, AST-1, PAG-3, CEH-16, EYG-1, CEH-45, MLS-2, CEH-34, LIM-7, CEH-41, TAB-1, Y56A3A.28, BAZ-2, HAM-2, UNC-39, PHA-4, TBX-2, CEH-27, Y75B8A.6, SOX-2, CRH-2, CES-2
Based on ChIP-seq (directly from ModERN database, targets from L1 stage ChIP-seq data are underlined)
Targeting: no-data
Targeted by: NA |