WormBase ID: WBGene00005009
WormBase description: Is an ortholog of human ZFAT (zinc finger and AT-hook domain containing). Is predicted to have DNA binding activity and metal ion binding activity. Is involved in response to oxidative stress. Is expressed in neurons. Is used to study Alzheimer's disease.
|
|
KI information: mNeonGreen knockin at N-terminus
sgRNA1: CAATCGAGTCGGACAATCTA
sgRNA2: GACTTTGTCGCTGCTGCCCA
5'HR: GCATGCACCAAGTTTCTGGAGTAGGGATTTTAAAACTGCCAGGGTTCGTTTGGAATTACTGTAAATTTCCTATCCTGCTATCGTTTTTTCAGCTAAACTTTTTTTCTTGAAGATATAAGTTAACCAACTGAAACATAGAGCATTTTGATGGGAAATTTGTGATTGCAAAACTTTAGAATTTATTTTTGAAAAGTCAGCATCTACTTGGAAGAACAACAATATAACTGACCTATCTCAAATCACTGTGTTACTTCAACTGCATTTCCTATAAAATTTAAATGATAGTGAGATATAATACAACGTAGATACTCCCTAAAAGGCCATTCCATAATGCCTTTTTTCTTTTTGCAACCATTATTTTTTCTTTCATCGGTCCCTCTTCCCTGTCTAATCCTTATTGTCTTACACACTCTCTCGGATGTTCTCTCTGTCTCTTCTTATAATCAAGTTTCGATCGATATTCACTAGCTGTTGTGAGGTTAGCGTTTTAGAAGCTGCACCAGAAAACCGAAAAGAGTGTTGGAGACGAAGATACGCAGATGGTCCGCCTATGAAAATTCTTGATTCATAGCGTCTCCTACCTTGGAATGAATTTATATTGTTAATCTATCCGCTTTACCTTTTTCTGGAACATTTAACATAAATGTTCGTTTATAGGAGAATATCACAAATCTCTCTTCACTCAAACCTCCACTTTCTGACTGTACGAGTCAAGATCACCGAATCAAGATCACCAATGTACTTTCCAGAAATTAAGCGTTTTGTATCGATTCTACATAAAATGAGACTTACCATTTTTAATTAATTAGTTTTTTCTCATATATTCGCTTGTTCTCTTTATTTTTCAAAAAAAAAAATTGAATTTTAATATAAATTTAGAAATCTAAACACAAAAAATTTCAGGGTAAGGTACC
3'HR: CTGCAGATGTCGTCGGAACCAACTTCTTCAATCGAGTCGGACAACCTACGACGCTCGAAACGAAAGAAGTTCAAGCTCGACTTCGTTGCCGCCGCTCATGGCAACAACCAGAAAAAGAGTCGTAAGGATCCTCATGGGCAGGGAGATGATGATGACGACACTTTTGATGACGATCTGAATCATTTTGAGCCATTGAGCGTTCTCACGGAGCAAGACGTTTCGATGGAAATGTTTGAAGATGATGAAGAGGATACTGTAAGCCGACGGAGGACGAGACGAAGCACGGCTCATTTTCAGGATTATCAAGAGCCTGACAGATGGATTGAGGTTAGAATCTTAAATTTCAATTTTTTTCAAACAATTTTTGCAGAACTCTGGCCCACATGCTTGCCATAAATGTCCATCCCGGTACGAGTCGAAAAGCAGTCTGGCGAACCACACAAAAATGCATCTTGGCGAGAAGCGGAAGTTTGCCTGTGAGCTGTGTGATTTCTCGGCGAGCACACTGAAATCTCTCACTCATCATAACAATATTCATCAGAATTTTGGGGTTCTTAGTCAGCAGACGAGTCCCGTGATTCCCGTGGCAAGCTCGGCAGATTCTACACTCAATTCTTCTATAAACTCCACTGGATGCCAAATCAATGCTCCACCTGCTCCGAAGCTCGACGAGGAAGCTCCAATTGCCGTAGAATCGGTAGTAGTTCATCATGAAGACGTTGCCGAATTTGACACTACACCTCCACCTATACTAGAACGTGAGGATGATGGACCTCCGGTATTGATAAGAGAAGCACCGGTTCGTAAATCGAATCGTCCAATGAAACCAACTCAAAAAGCTCAAAAAATGACAAAGCAACGAGAGCGGAAGTCGAAAACAGTTGAATCACCGAAAGGATCACTTCCATCATCATCCGCATCTTCAGTAACTCCACCACCAGTGAGAAAAGATGTCGAAAAAACTAGT
KI verification primers: SPR-4-NKIF: 5'-TTTACCACAATTTTTCAACTGGGTT-3' SPR-4-NKIR: 5'-GTAGTGAAAGGACAATGAGGGCA-3' Download
|
Strain name: SYS475 KI background: JIM113 Strain genotype: ujIs113 (pie-1p::mCherry::H2B; nhr-2p::mCherry::his-24::let858 3'UTR; unc-119(+)) II; spr-4 (devKi127 [mNeonGreen::spr-4]) I
|
Expression pattern: Ubiquitous
|
Lineage expression pattern
350 stage
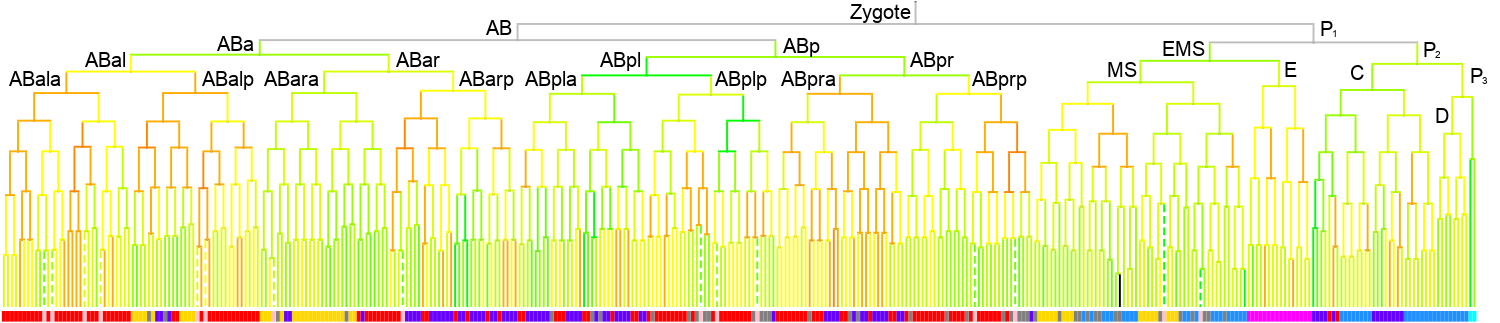
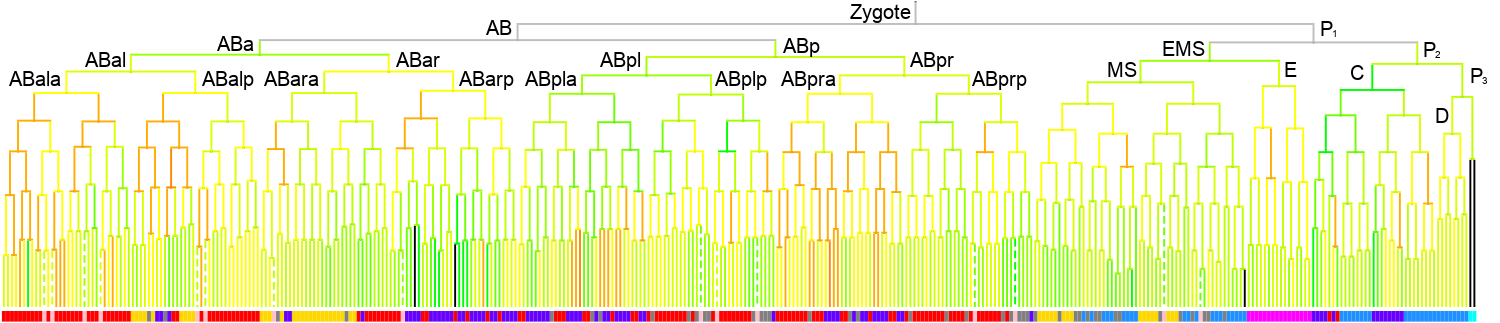
|
600 stage
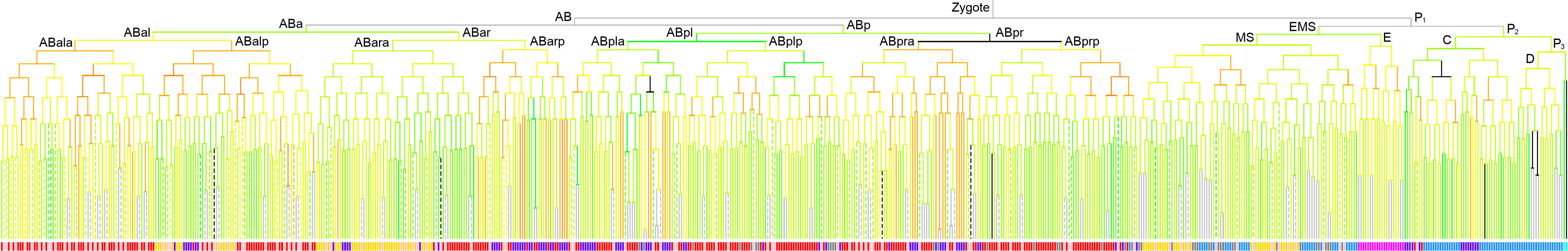 |
3D expression pattern  4-cells  8-cells  15-cells  28-cells  ~50-cells  ~100-cells  ~200-cells  ~350-cells  ~500-cells  ~600-cells  600-cells  bean-stage |
Inferred TF relationships
Based on expression similarity (strong, medium, weak): NA
Based on ChIP-seq (directly from ModERN database, targets from L1 stage ChIP-seq data are underlined)
Targeting: no-data
Targeted by: ALY-1, CEH-36, CEH-39, CEH-43, CEH-48, MLS-2, NFYA-1, PHA-2, UNC-120, UNC-42, UNC-55, CEH-82, DPL-1, EFL-1, LSY-2, MEP-1, RNT-1, UNC-86 |
|
 Based on reannotated ChIP-seq (peaks are assigned to a target if it is located within 2 kb relative to the transcription start site; targets from L1 stage ChIP-seq data are underlined) Based on reannotated ChIP-seq (peaks are assigned to a target if it is located within 2 kb relative to the transcription start site; targets from L1 stage ChIP-seq data are underlined)
Targeting: no-data
Targeted by: CEH-39, CEH-48, NFYA-1, UNC-42, CEH-82, EFL-1, LSY-2, RNT-1, UNC-86
|