WormBase ID: WBGene00009133 WormBase description: Is an ortholog of human ZBED1 (zinc finger BED-type containing 1); ZBED4 (zinc finger BED-type containing 4); and ZBED6 (zinc finger BED-type containing 6). Exhibits RNA polymerase II regulatory region sequence-specific DNA binding activity. Is involved in molting cycle; regulation of cell division; and vulval cell fate specification. Is expressed in M lineage cell; hypodermal cell; and vulval cell. |
Reporter verification: Verified by PCR and sequencing |
Strain name: SYS563 Cross: OP651 x JIM113 Strain genotype: ujIs113 (pie-1p::mCherry::H2B; nhr-2p::mCherry::his-24::let858 3'UTR; unc-119(+)) II; wgIs651 (bed-3::TY1::EGFP::3xFLAG + unc-119(+)) |
Expression pattern: Non-specific |
Lineage expression pattern 350 stage   |
600 stage 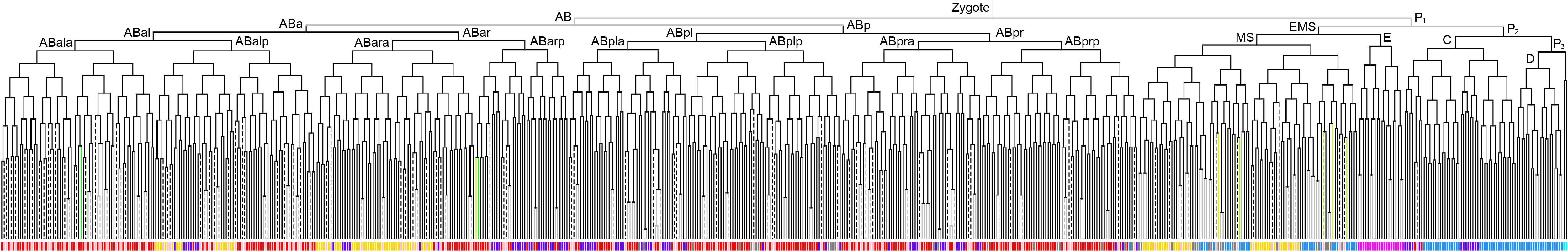 |
3D expression pattern  4-cells  8-cells  15-cells  28-cells  ~50-cells  ~100-cells  ~200-cells  ~350-cells  ~500-cells  ~600-cells  600-cells  bean-stage |
Inferred TF relationships Based on expression similarity (strong, medium, weak): CEH-31, UNC-39, UNC-120, F23B12.7 Based on ChIP-seq (directly from ModERN database, targets from L1 stage ChIP-seq data are underlined) Targeting: AST-1, CEH-2, DAF-19, DMD-5, ETS-5, K09A11.1, LIM-7, MDL-1, NHR-102, RCOR-1 |
|
|