WormBase ID: WBGene00003847 WormBase description: Is an ortholog of human PRDM1 (PR/SET domain 1). Exhibits proximal promoter sequence-specific DNA binding activity. Is involved in several processes, including dauer entry; negative regulation of distal tip cell migration; and regulation of transcription by RNA polymerase II. Localizes to cytoplasm and nucleus. Is expressed in several structures, including distal tip cell; dorsal nerve cord; enteric muscle; neurons; and tail hypodermis. |
Reporter verification: Verified by PCR and sequencing |
Strain name: SYS458 Cross: OP109 x JIM113 Strain genotype: ujIs113 (pie-1p::mCherry::H2B; nhr-2p::mCherry::his-24::let858 3'UTR; unc-119(+)) II; wgIs109 (blmp-1::TY1::EGFP::3xFLAG + unc-119(+)) |
Expression pattern: Skin-specific |
Lineage expression pattern 350 stage   |
600 stage 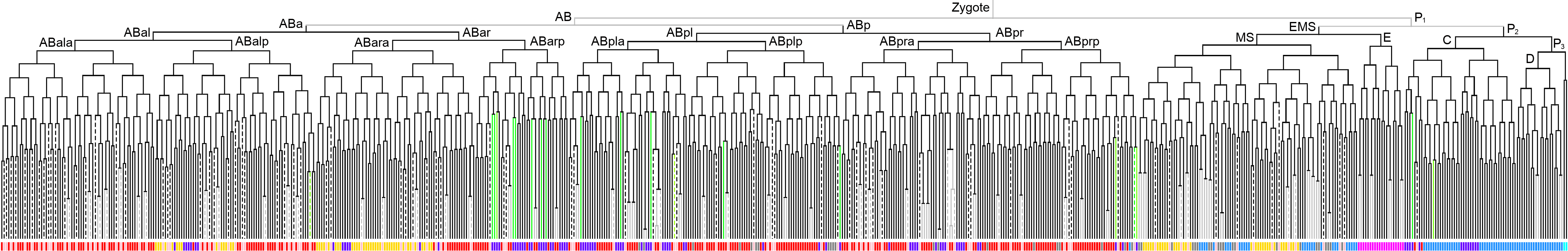 |
3D expression pattern  4-cells  8-cells  15-cells  28-cells  ~50-cells  ~100-cells  ~200-cells  ~350-cells  ~500-cells  ~600-cells  600-cells  bean-stage |
Inferred TF relationships Based on expression similarity (strong, medium, weak): NHR-23, LIN-26, HAM-2, NHR-25, CEH-40, ELT-1, HBL-1, CEH-20, Y75B8A.6, F23B12.7, CEH-43, CRH-2 Based on ChIP-seq (directly from ModERN database, targets from L1 stage ChIP-seq data are underlined) Targeting: NHR-25, ATFS-1, BCL-11, DAF-12, DVE-1, EFL-3, ETS-4, FOS-1, HLH-30, LAG-1, LIN-15B, MDL-1, MEP-1, RNT-1, SEM-2, SKN-1, UNC-130, UNC-4, UNC-55, UNC-62, Y55F3AM.14, ZAG-1, ZTF-30, NHR-23, HAM-2, ELT-1, HBL-1, CEH-20, Y75B8A.6, CEH-43, CRH-2, ALR-1, ALY-1, ATF-7, ATHP-1, BED-3, CAMT-1, CEBP-1, CEH-13, CEH-14, CEH-16, CEH-18, CEH-38, CEH-48, CEH-88, CFI-1, DAF-19, DAF-3, DNJ-17, EFL-1, EGL-18, EGL-44, EGRH-1, ELT-3, EOR-1, ETS-5, FKH-6, GEI-3, GMEB-1, GMEB-2, HLH-11, HLH-14, HLH-2, HMBX-1, IRX-1, KLF-3, LIM-7, LIN-1, LIN-11, LIN-13, LIN-22, LIN-39, LSY-2, MEF-2, MML-1, NFI-1, NFYA-1, NGN-1, NHR-10, NHR-12, NHR-127, NHR-35, NHR-49, NHR-76, NHR-85, NOB-1, PHA-4, POP-1, PROS-1, RCOR-1, SDZ-38, SEL-8, SEM-4, SMA-2, SWSN-7, T07F8.4, TLP-1, TTX-1, UNC-3, UNC-30, UNC-37, VAB-15, VAB-3, Y73E7A.1, ZFH-2, ZIP-3, ZTF-11, ZTF-6 Targeted by: HAM-2, ELT-1, BCL-11, CEH-31, CEH-36, ELT-4, HMG-4, LIN-32, LSY-2, NFYA-1, PROS-1, RNT-1, SPTF-1, UNC-62, ALY-2, C16A3.4, CEH-82, DPL-1, EFL-1, ELT-2, ELT-3, MEP-1, NHR-85, PQM-1, SWSN-7 |
|
Targeting: NHR-25, ATFS-1, DVE-1, EFL-3, ETS-4, LAG-1, MDL-1, SEM-2, SKN-1, UNC-4, UNC-55, UNC-62, ZAG-1, ZTF-30, NHR-23, HAM-2, ELT-1, HBL-1, CEH-20, CRH-2, ALR-1, ALY-1, ATF-7, ATHP-1, BCL-11, BED-3, CEBP-1, CEH-13, CEH-16, CEH-48, CEH-5, CEH-88, DAF-19, DPL-1, EFL-1, EGL-18, EGL-44, ELT-3, EOR-1, ETS-5, FOS-1, GEI-3, GMEB-1, HLH-30, HMBX-1, IRX-1, K10B3.5, KLF-3, LIN-1, LIN-13, LIN-15B, LIN-39, LSY-2, MEF-2, MEP-1, MML-1, NFI-1, NFYA-1, NFYC-1, NHR-10, NHR-12, NHR-127, NHR-35, NHR-49, NHR-76, NHR-85, NOB-1, POP-1, PROS-1, RCOR-1, RNT-1, SEL-8, SEM-4, SMA-2, SVH-5, SWSN-7, T07F8.4, TLP-1, UNC-130, UNC-37, VAB-15, Y73E7A.1, ZIP-3 Targeted by: HAM-2, BCL-11, CEH-31, ELT-4, HMG-4, LSY-2, NFYA-1, PROS-1, SPTF-1, ELT-2, ELT-3, MEP-1, PQM-1, SWSN-7 |