WormBase ID: WBGene00007772 WormBase description: Is an ortholog of human EGR1 (early growth response 1); EGR2 (early growth response 2); and EGR3 (early growth response 3). Is predicted to have DNA-binding transcription factor activity, RNA polymerase II-specific. Is involved in negative regulation of MAP kinase activity; negative regulation of oocyte maturation; and negative regulation of ovulation. Localizes to nucleus and perinuclear region of cytoplasm. Is expressed in body wall musculature; hermaphrodite somatic gonadal cell; nervous system; pharynx; and sperm. Human ortholog(s) of this gene are implicated in several diseases, including Charcot-Marie-Tooth disease (multiple); Denys-Drash syndrome; and Frasier syndrome. |
Strain name: SYS444 KI background: JIM113 Strain genotype: ujIs113 (pie-1p::mCherry::H2B; nhr-2p::mCherry::his-24::let858 3'UTR; unc-119(+)) II; egrh-1 (devKi113 [mNeonGreen::egrh-1]) X |
Expression pattern: Non-specific |
Lineage expression pattern 350 stage   |
600 stage 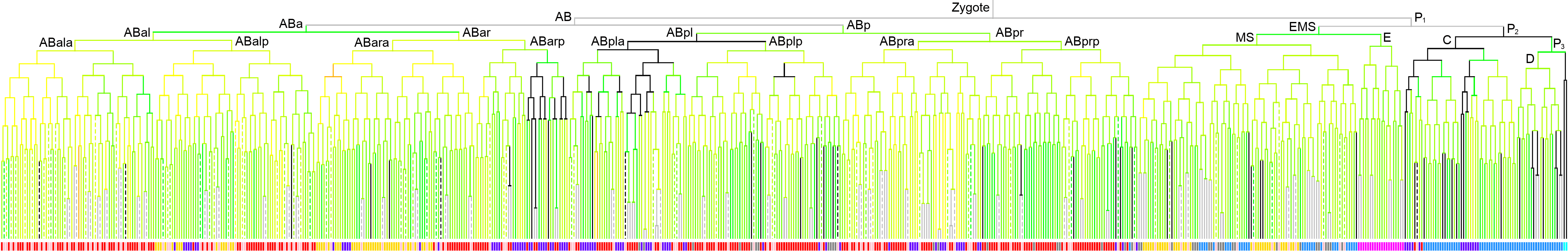 |
3D expression pattern  4-cells  8-cells  15-cells  28-cells  ~50-cells  ~100-cells  ~200-cells  ~350-cells  ~500-cells  ~600-cells  600-cells  bean-stage |
Inferred TF relationships Based on expression similarity (strong, medium, weak): NA Based on ChIP-seq (directly from ModERN database, targets from L1 stage ChIP-seq data are underlined) Targeting: no-data Targeted by: BCL-11, CEH-14, CEH-18, CEH-19, CEH-2, CEH-31, CEH-34, CEH-39, CND-1, COG-1, DMD-4, DVE-1, ELT-2, FKH-6, HLH-1, HLH-30, HLH-8, HMG-4, LIM-6, LSY-2, MEP-1, NFYA-1, PAG-3, PHA-2, PHA-4, PROS-1, REF-2, RNT-1, SYD-9, TBX-2, UNC-120, UNC-3, UNC-42, UNC-55, UNC-62, ALY-2, BLMP-1, C16A3.4, CEH-8, CEH-82, DPL-1, DSC-1, EFL-1, EGL-13, ELT-3, FKH-10, FKH-8, FOS-1, IRX-1, LIN-13, LIN-39, MDL-1, MEF-2, PAX-3, SKN-1, SOX-4, SWSN-7, UNC-130, UNC-86, ZAG-1, ZTF-11 |
|
Targeting: no-data |