WormBase ID: WBGene00003611 WormBase description: Is predicted to have DNA-binding transcription factor activity. |
Reporter verification: Whole-genome sequencing indicated that the GFP tags another TF, NHR-12 rather than CEH-30. |
Strain name: SYS93 Cross: OP120 x JIM113 Strain genotype: ujIs113 (pie-1p::mCherry::H2B; nhr-2p::mCherry::his-24::let858 3'UTR; unc-119(+)) II; wgIs120 (nhr-12::TY1::EGFP::3xFLAG + unc-119(+)) |
Expression pattern: Lineage-specific, Tissue-specific (Skin and Intestine), Transient-expression |
Lineage expression pattern 350 stage 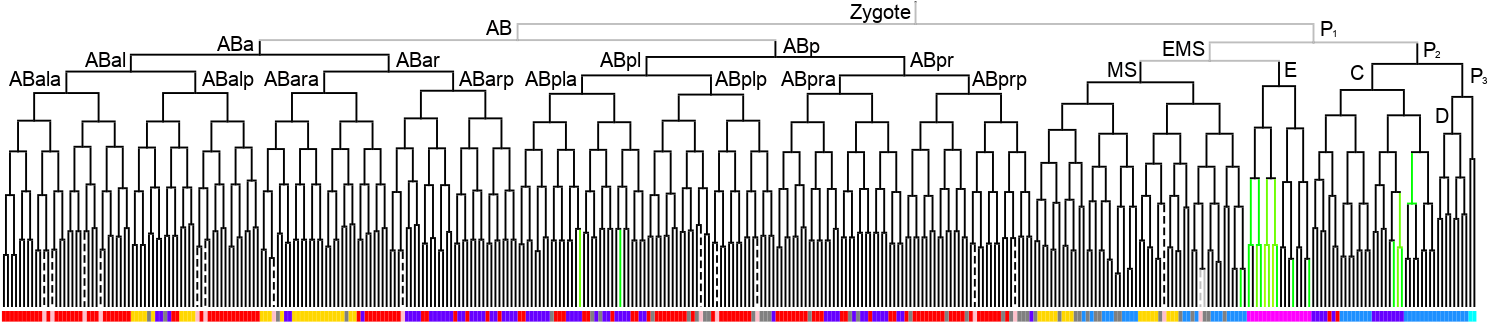 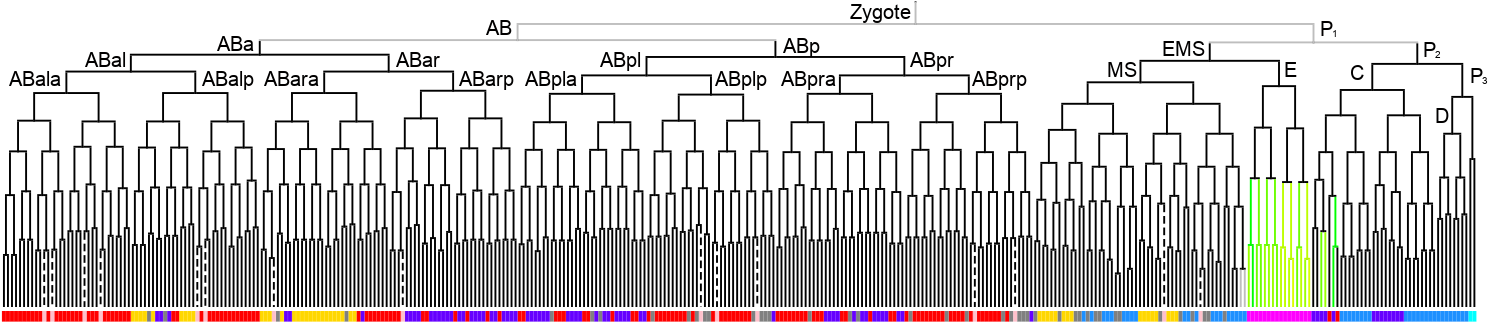 |
600 stage 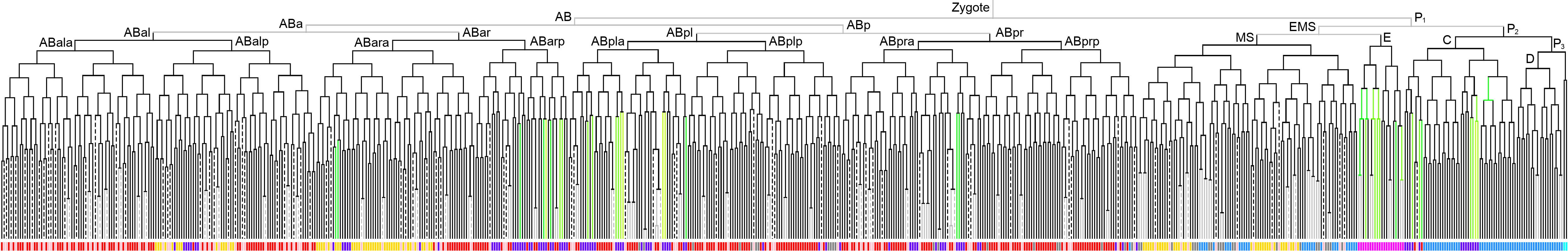 |
3D expression pattern  4-cells  8-cells  15-cells  28-cells  ~50-cells  ~100-cells  ~200-cells  ~350-cells  ~500-cells  ~600-cells  600-cells  bean-stage |
Inferred TF relationships Based on expression similarity (strong, medium, weak): C16A3.4, MDL-1, ELT-3, ELT-2, PQM-1, ELT-7, END-1, RNT-1, NHR-102, NHR-232, GMEB-1, NHR-23, CEH-16, NOB-1, LIN-26, CEH-40, REF-1, DVE-1, BAZ-2, HAM-2, ELT-1, NHR-25, CEH-20, HBL-1, VAB-7, TBX-9, ATF-7, TLP-1, TBX-11, UNC-62, TBX-8, F23B12.7, CEH-74, NFI-1, SEL-8, CEBP-1, LAG-1, Y75B8A.6, NHR-10, CRH-2 Based on ChIP-seq (directly from ModERN database, targets from L1 stage ChIP-seq data are underlined) |
|
|