WormBase ID: WBGene00001949 WormBase description: Is an ortholog of human TCF12 (transcription factor 12); TCF3 (transcription factor 3); and TCF4 (transcription factor 4). Exhibits several functions, including DNA-binding transcription factor activity, RNA polymerase II-specific; bHLH transcription factor binding activity; and protein dimerization activity. Contributes to RNA polymerase II regulatory region sequence-specific DNA binding activity. Is involved in several processes, including generation of neurons; gonad morphogenesis; and nematode male tail tip morphogenesis. Localizes to RNA polymerase II transcription factor complex and cytoplasm. Is expressed in several structures, including distal tip cell; lateral ganglion; non-striated muscle; ray precursor cell; and terminal bulb. Human ortholog(s) of this gene are implicated in Fuchs' endothelial dystrophy; Pitt-Hopkins syndrome; and craniosynostosis. |
Reporter verification: Verified by comparing expression pattern to the literature (Krause et al., 1997) |
Strain name: SYS1049 Cross: OH12412 x JIM113 Strain genotype: ujIs113 (pie-1p::mCherry::H2B; nhr-2p::mCherry::his-24::let858 3'UTR; unc-119(+)) II; otIs502 (hlh-2(fosmid)::YFP + myo-3p::mCherry) |
Expression pattern: Lineage-specific, Neuronal-specific, Transient-expression |
Lineage expression pattern 350 stage   |
600 stage 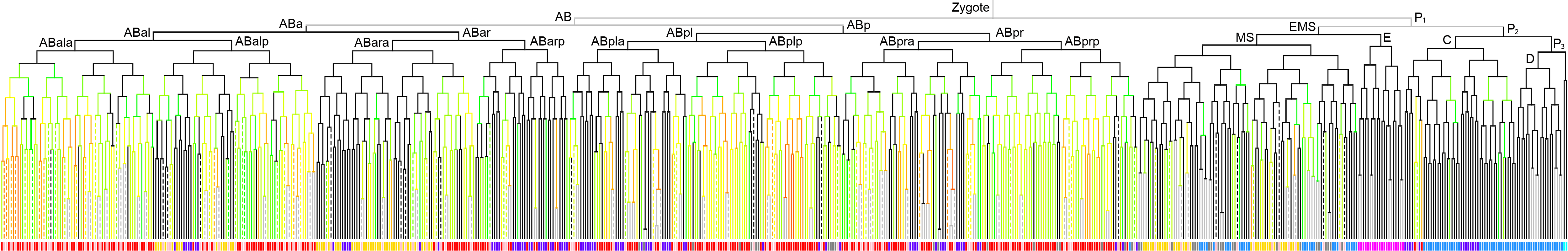 |
3D expression pattern  4-cells  8-cells  15-cells  28-cells  ~50-cells  ~100-cells  ~200-cells  ~350-cells  ~500-cells  ~600-cells  600-cells  bean-stage |
Inferred TF relationships Based on expression similarity (strong, medium, weak): CEH-10, TTX-3, UNC-3, HLH-14, CND-1, LIN-32, ZAG-1, UNC-42, NGN-1, CEH-48, UNC-86, LIN-11, CEH-31, TAB-1, HLH-3, UNC-30, CEH-32, EGL-46, PROS-1, MBF-1, EGL-44, NHR-67, HLH-30, VAB-3, ZTF-11, CEH-30, COG-1, SOX-2 Based on ChIP-seq (directly from ModERN database, targets from L1 stage ChIP-seq data are underlined) Targeting: no-data Targeted by: CND-1, UNC-42, CEH-31, HLH-30, COG-1, CEH-34, CEH-36, CEH-39, ELT-1, GMEB-2, HAM-2, HLH-8, HMG-4, LIN-13, M03D4.4, MLS-2, NFYA-1, PHA-2, PHA-4, REF-2, SPTF-1, UNC-62, ZIP-8, BLMP-1, DPL-1, DSC-1, EFL-1, FKH-4, FKH-8, LSY-2, MEP-1 |
|
Targeting: no-data Targeted by: CND-1, UNC-42, CEH-31, HLH-30, COG-1, CEH-34, CEH-36, CEH-39, ELT-1, GMEB-2, HAM-2, HMG-4, LIN-13, M03D4.4, MLS-2, NFYA-1, PHA-2, PHA-4, REF-2, SPTF-1, EFL-1, FKH-8 |