WormBase ID: WBGene00003018
WormBase description: Is an ortholog of human ATOH1 (atonal bHLH transcription factor 1). Exhibits protein heterodimerization activity; protein homodimerization activity; and sequence-specific DNA binding activity. Is involved in several processes, including embryo development; nematode male tail tip morphogenesis; and neuron development. Localizes to nucleus. Is expressed in several structures, including QL.a; QL.p; QR.a; oxygen sensory neurons; and ray precursor cell. Human ortholog(s) of this gene are implicated in congenital malabsorptive diarrhea 4 and persistent hyperplastic primary vitreous.
|
|
KI information: mNeonGreen knockin at N-terminus
sgRNA1: GGCACTGTGGCACATACATT
sgRNA2: TGGTGGACTGAATTGATCCT
5'HR: GCATGCCTGAAATTTTTAGATTTTTTTTTCAAAGTTTTCATTCAAGAGTTTCCCAGTTTTCCAGAAAAAATTGGACTAACTAGATAAATGTTGGAGGAGTATCTAGAATTTAAGAATAAGAACAATTGAAACGCGATTGAGTTTCATAATTATGACCATTTTTTGGACAAAAACCAAAAATTTCCAAACATTTTTTTAAACGTTTCGCTGGGAAACTTAAATATTTAGCACGAAAATTGAAATTCTTACGGCGTTTGTATTTCATATTTCCTGAACACCGACTAGATTTAATTATTAGAATTTGTTTTTTTTTCCTTATTTTCTGGTTATAAATTACGCTAATATACGTTTATAAAATCACGAATTTTATAAAATCAAGCTTGCTTAGATTTTTTTTAAAAAGTTTTCTAAATTCTTCAGTCTTGTAGGCTAAATATTTTATTTCCATCATTTTTATAAGTACTTGTGTAGTGTGCCTAGCCGAAAAATATTTTCCAATTTGTTTTCCAATTATATCAATTATATGGATAATTTAAGTGAAATTGATTTGTATTCCTTGGCTTAGAGAAACACTAATTTTAAGCCTAAAACTATAGATAAAAACGTCACGTAAAATGTTCAAAATGGTAAATTTGATAATTCTGTGCTGGTATACTACACAAATACCGTTTCTCTATATTTTCAAGTTTAAGCTCAAGAGCTTCACTGTTATTGCAAGTTCAAACTCAACTTAGCATTTTTCAACCAAACCGTTTCCATTTGGTTTTTGTAACCCTTCTTACACGCTCTTTCACAATGGCAGATAATTAATCACCTTGCCTCCTCCCAGTCGTCTCCGCACACGCCTCCTCCATATTAGAGTACTGTAGATGCCGGGTGGGCGGAGCTTCGTCTTCTGTTTCTTTGCATCGAATCAACAGGTTATTCTCTCACACATCGTCGTTTTCAGTCAGACCAACCGGTACC
3'HR: CTGCAGATGAGCTGGGAGCAATATCAAATGTATGTGCCACAGTGCCATCCAAGCTTCATGTATCAAGGATCAATTCAGTCCACCATGACAACTCCACTGCAAAGCCCCAACTTTTCACTCGATTCTCCAAATTACCCGGATAGTCTTTCGTAAGTTTAACCATATCATCTTTTTTTCATTAAAATATTGATTTCTAATTGATTTCTTGTTGCAGAAATGGAGGAGGAAAAGATGATAAAAAGAAGTGCCGCCGTTATAAGACACCGTCCCCACAATTGCTCCGAATGAGAAGAAGTGCTGCAAATGAACGGGAGCGCCGAAGAATGAACACGCTGAATGTTGCCTATGACGAGGTTAGTTTATTCTTGAATTGTAATCGTTCAATTCAATTATCTGAAGAAGTAGCTCGAGCCAAAACATTTAAGTACATGCTTTATATTTTGTGAATAATTCAGTATTGTTTTTGGAAAATCATTTGTCATAGTCATCAGTAGTCACTCAAAAATTCAAAATCATAACCGATATATAGTAGTATACTGTAAGATGTGCTGTCATGAAGGACATGTATGCTAACAGATGTTATCCAACCACTACTCGAGTCGTTTGCGTTTACCTCACTCACGACTACACCAATTCTCAATTCTCAATTTCAATTCCCACCACTCAATTTATGACTGACCATTTCAAATTTATGCCAAATTAAATGTGGCCGTTGCAAAAATTCGTTACTATTTCAGCTTCGCGAGGTTCTTCCTGAAATCGACTCGGGGAAGAAGTTGTCCAAATTCGAAACATTGCAAATGGCTCAGAAGTATATTGAATGCCTGTCGCAGATTTTGAAGCAAGACTCCAAAAATGAGAATCTCAAAAGCAAATCCGGATGATCATCTATTTTTTAAAATTTTTTTTTCAATCTTTCATTCGTCGTGTCTTTATCATTACTAGT
KI verification primers: LIN-32-NKIF: 5'-TTTTGATAAGGATCTAATGAAGTGTGAA-3' LIN-32-NKIR: 5'-ACTAAAGCACCCATAATCGAGGA-3' Download
|
Strain name: SYS398 KI background: JIM113 Strain genotype: ujIs113 (pie-1p::mCherry::H2B; nhr-2p::mCherry::his-24::let858 3'UTR; unc-119(+)) II; lin-32 (devKi70 [mNeonGreen::lin-32]) X
|
Expression pattern: Lineage-specific, Neuronal-specific
|
Lineage expression pattern
350 stage
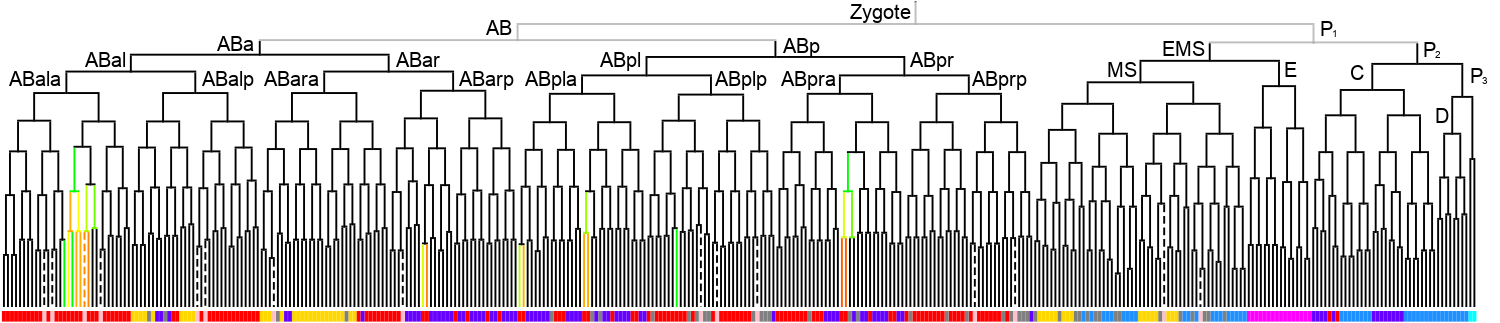

|
600 stage
 |
3D expression pattern  4-cells 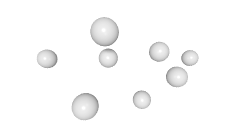 8-cells  15-cells  28-cells  ~50-cells 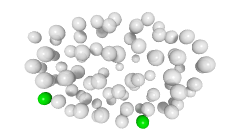 ~100-cells 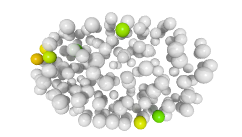 ~200-cells  ~350-cells  ~500-cells  ~600-cells  600-cells  bean-stage |
Inferred TF relationships
Based on expression similarity (strong, medium, weak): CEH-10, CEH-31, UNC-86, NOB-1, CEH-30, Y73E7A.1, UNC-39, VAB-3, CEH-32, EGL-46, Y56A3A.28, EFL-3, HLH-30, CEH-43, HLH-2, SEM-2, EGL-44, TBX-2, SPTF-1, ZTF-11, HLH-3, CES-2, SOX-2, ELT-6, ELT-1, ZIP-8
Based on ChIP-seq (directly from ModERN database, targets from L1 stage ChIP-seq data are underlined)
Targeting: UNC-39, EGL-46, EFL-3, HLH-30, CEH-43, EGL-44, ZTF-11, CES-2, AHR-1, ALY-1, AST-1, BLMP-1, CEH-14, CEH-2, CEH-20, CEH-39, DAF-19, DMD-5, EGL-18, ETS-5, HBL-1, K09A11.1, LIM-7, NHR-102, POP-1, SEL-8, SWSN-7, SYD-9, TTX-3, UNC-42, UNC-62, ZAG-1, ZTF-6
Targeted by: ELT-1, ZIP-8, ELT-2, TBX-2, ZTF-11 |
|
 Based on reannotated ChIP-seq (peaks are assigned to a target if it is located within 2 kb relative to the transcription start site; targets from L1 stage ChIP-seq data are underlined) Based on reannotated ChIP-seq (peaks are assigned to a target if it is located within 2 kb relative to the transcription start site; targets from L1 stage ChIP-seq data are underlined)
Targeting: EGL-46, HLH-30, EGL-44, ZTF-11, CES-2, AHR-1, ALY-1, BAZ-2, CEH-14, CEH-2, CEH-20, CEH-39, DAF-19, DMD-5, ETS-5, HBL-1, SWSN-7, SYD-9, TTX-3, UNC-42, UNC-62, ZTF-6
Targeted by: ELT-1, ZIP-8, ELT-2, TBX-2, ZTF-11
|














