WormBase ID: WBGene00003595 WormBase description: Is an ortholog of human NEUROG1 (neurogenin 1) and NEUROG3 (neurogenin 3). Is predicted to have protein dimerization activity. Is expressed in ABaraappaa. Human ortholog(s) of this gene are implicated in congenital malabsorptive diarrhea 4; diabetes mellitus (multiple); and persistent hyperplastic primary vitreous. |
Strain name: SYS498 KI background: JIM113 Strain genotype: ujIs113 (pie-1p::mCherry::H2B; nhr-2p::mCherry::his-24::let858 3'UTR; unc-119(+)) II; ngn-1 (devKi137 [mNeonGreen::ngn-1]) IV |
Expression pattern: Lineage-specific, Neuronal-specific, Transient-expression |
Lineage expression pattern 350 stage 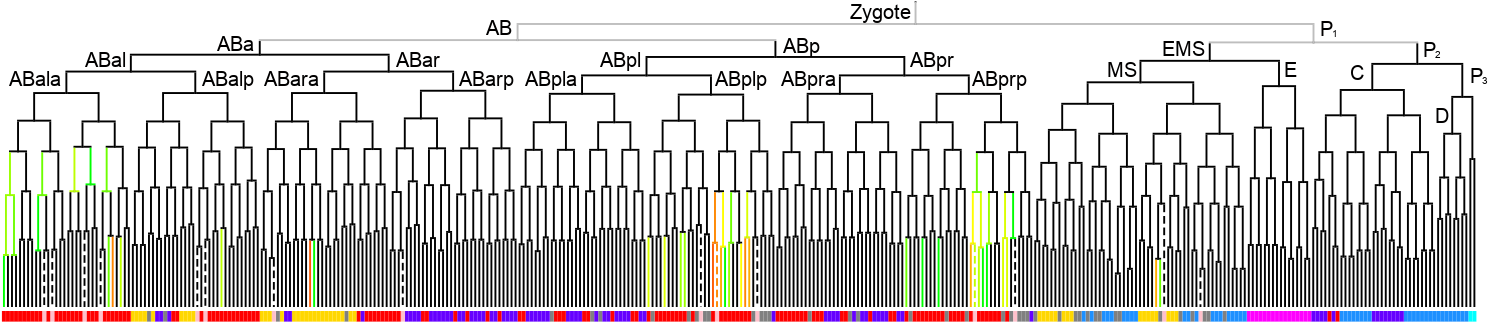 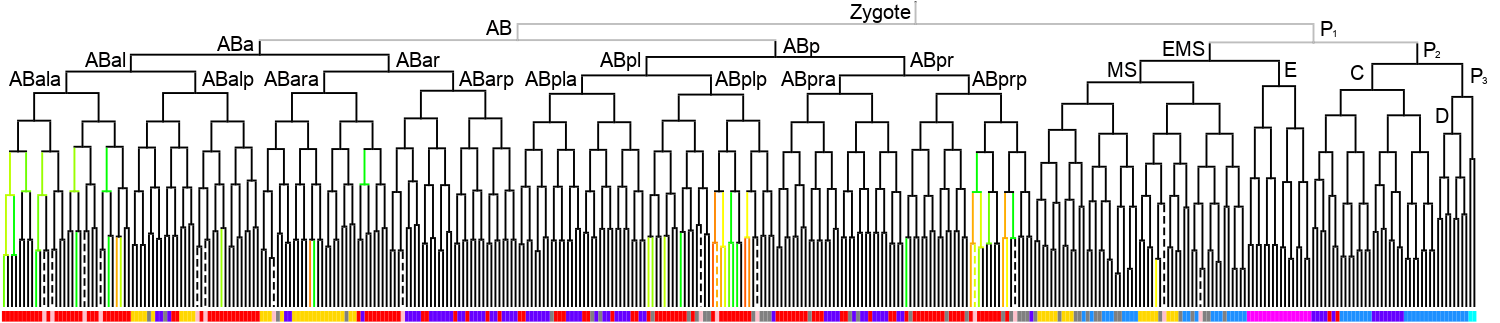 |
600 stage 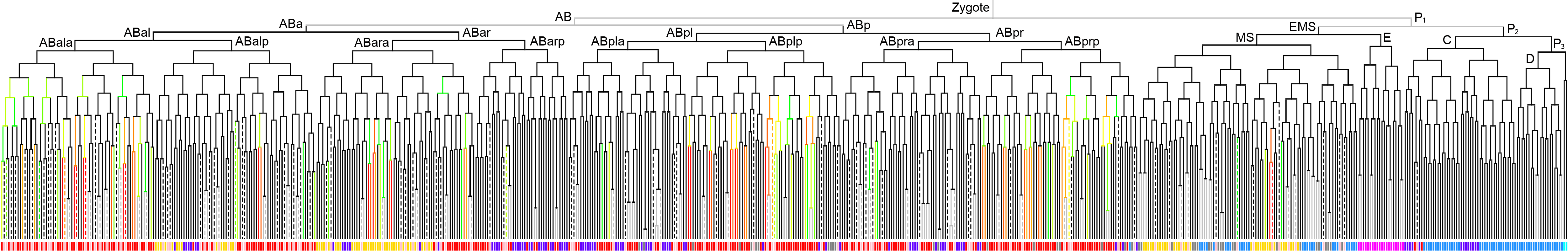 |
3D expression pattern  4-cells  8-cells 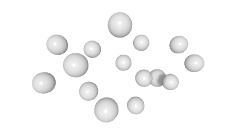 15-cells  28-cells  ~50-cells 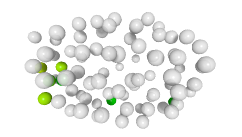 ~100-cells  ~200-cells  ~350-cells  ~500-cells  ~600-cells 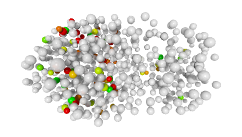 600-cells  bean-stage |
Inferred TF relationships Based on expression similarity (strong, medium, weak): AHA-1, FKH-10, CEH-17, LIM-4, UNC-42, EGL-13, EGL-46, CEH-10, CEH-48, ZAG-1, TAB-1, EGL-44, TTX-1, MLS-2, UNC-30, HLH-2, HLH-3, CEH-30, CEH-27, ZTF-11, VAB-3, HLH-30, ELT-6, CEH-36, SOX-2, CEH-32 Based on ChIP-seq (directly from ModERN database, targets from L1 stage ChIP-seq data are underlined) Targeting: no-data Targeted by: CEH-36, ELT-2, LSY-2, UNC-62, ZIP-8, BLMP-1, DPL-1, EFL-1 |
|
Targeting: no-data Targeted by: ELT-2 |