WormBase ID: WBGene00003912 WormBase description: Is an ortholog of human CDX1 (caudal type homeobox 1). Exhibits DNA-binding transcription factor activity, RNA polymerase II-specific and RNA polymerase II regulatory region sequence-specific DNA binding activity. Is involved in several processes, including ectodermal cell fate commitment; muscle cell fate commitment; and positive regulation of transcription by RNA polymerase II. Localizes to condensed nuclear chromosome. Is expressed in several structures, including ABprpppppaa; enteric muscle; oocyte; rectal epithelial cell; and tail spike. |
Reporter verification: Verified by PCR and sequencing |
Strain name: RW10993 Strain genotype: unc-119 (ed3) III; itIs37 (pie-1p::mCherry::H2B::pie-1 3'UTR + unc-119(+)); stIs10116 (his-72p::his-24::mCherry::let-858 3'UTR + unc-119(+)); wgIs94 (pal-1::TY1::eGFP::3xFLAG(P000006_G02)) |
Expression pattern: Lineage-specific, Muscle-specific, Transient-expression |
Lineage expression pattern 350 stage  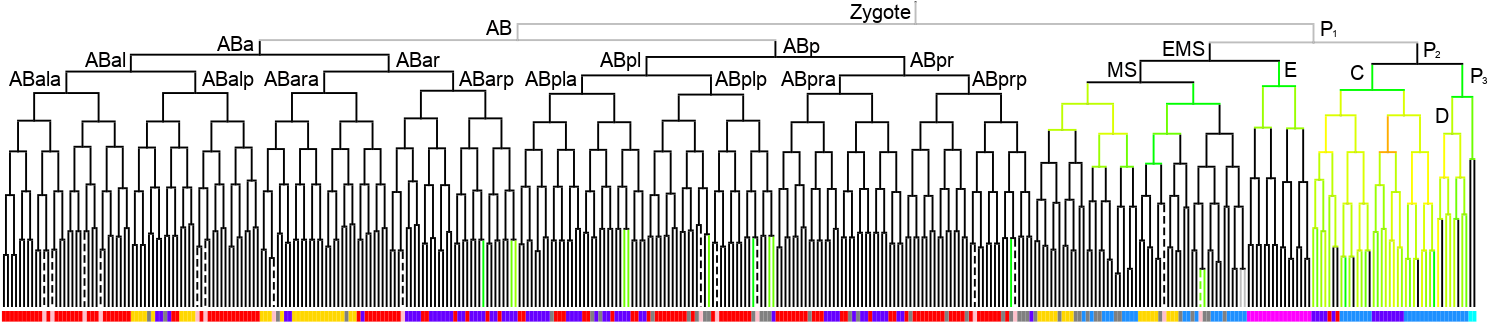 |
600 stage 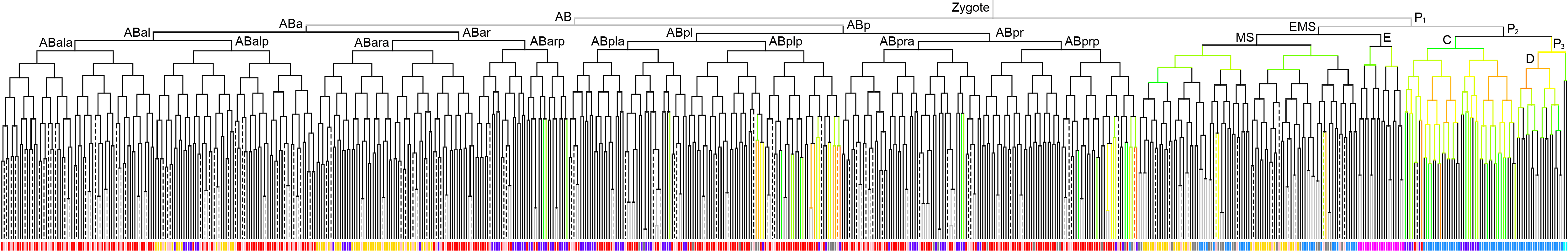 |
3D expression pattern  4-cells 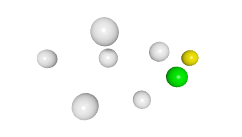 8-cells 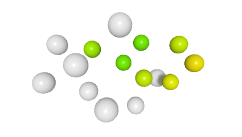 15-cells  28-cells  ~50-cells  ~100-cells  ~200-cells  ~350-cells  ~500-cells  ~600-cells  600-cells  bean-stage |
Inferred TF relationships Based on expression similarity (strong, medium, weak): ELT-3, LSL-1, HLH-8, VAB-7, SNAI-1, HND-1, EGL-5, EGL-38, MED-2, HLH-1, LIN-39, PHP-3, UNC-120, TBX-9, TLP-1, COG-1, HMBX-1, CEH-20, UNC-62, M03D4.4, HAM-2, LIN-26, NHR-25, HBL-1 Based on ChIP-seq (directly from ModERN database, targets from L1 stage ChIP-seq data are underlined) Targeting: no-data |
|
Targeting: no-data |