WormBase ID: WBGene00006462
WormBase description: Is an ortholog of human EHF (ETS homologous factor). Is predicted to have DNA-binding transcription factor activity, RNA polymerase II-specific. Is expressed in intestine and neurons. Human ortholog(s) of this gene are implicated in acute myeloid leukemia.
|
|
KI information: mNeonGreen knockin at C-terminus
sgRNA1: CTGGTGTACAAGTTCGGACC
sgRNA2: CCATGGTTGGGAAAATGTAA
5'HR: GCATGCTTTTCTCTTTGTTTCAGAAGACACAACTCTTGATACTGTCATTGGGCCATTTGTGGACGATGAAATTGCGACAACCAGTGCACCACCTACGATCGTTGATATGACTTCCTTGGGATTACAACCAATGATTAGCAGTATCACATGCACAATTCAGCCTCCTCTTCAACATCCAACGCCACACCTTGCTCCGCTACCTGTTATGCAACATCAGCTAAACAATTTGAGCAGTCTAACTCAAGTGCTCTCTCAACAGACATTTAACACTACAATTCTCACACCCCAGCAAACTGCCAACGCCGTTCAATCGCTCAATCAATCTTTGGCGTCCAGCATTGCAGCAGGCTTGACCAGTAGTCTCAGTCCATTAGGAGTCAACTATCAGCTATCAAGCCGGTTAAACCCCGCGTCACCGCTTCTCAGTGCGCCGATCCAACCAAAGTTGTCGCCAATGCTCAAATATTCATGCACGATCAACACCGAATACGAAGGTAAGAGCCAAATTCCAGACTCATGTTTTATTCAATATCCAATCTATTTTAGTGCCAGAAGAAAGGTCGGCTGAAAGCAGTGACTTGAAAATCAAAAAGAACAAAGATGGCAAACCAAGAAAGCGTTCGCAACACACGAAGGGTAATAAATTGTGGGAGTTTATCAGAGATGCGTTGAAAGACCCATCAACTTGTCCGTCTGTTGTGAGGTGAGTTCCTTCAGCAAATGTAAAAATAAAATTAAATACTCATGGTTAATGTTTCCAGATGGGAAGACCCGATTGAAGGAGTATTCCGTATCGTGGAGTCAGAGAAGCTGGCTCGGCTGTGGGGAGCTCGAAAAAACAATGAAAAAATGACATATGAAAAGCTCAGTCGAGCGATGAGGTTTGTCGTCAAAAATTCAAATTAATGCTTAAAAACATATCTTTTTTTCAGAACCTACTACGAAAAGCAAATACTGGTTCCAGTCCCAAAAACTGGTCTTTATCCGAAGAAACTCGTCTACAAATTTGGACCAGGTGCCCATGGTTGGGAAAACGTAAAAGGCATTGGTACC
3'HR: CTGCAGATGGTAAATTTGAATTTTTAAAGGAGCTAAATTAGATGTTATATACTTTTCAGGTTTCCAAGAAATTCCAGTCAATCAGTGTCAGGTATTGAATAGCACAGCAATGCTATTCATTGGAAGAATTCTAAACCAAACAGAAACTCAAATATACAAATCAAATTCCAGTTTTTTGATCTCTAATTGATGTATTCATTGGTAGTCGTATGGGAACACATTAACTTATGGTTTTTTTTTCGTCGTTATTAAAGTTTCCATATTTAAAAATATCTTATCTTCTCTCATCATCATCATCATTCAGTTTTCCCCGAAAAATTTGAGTTTTGTGATTTATAAATTTTGTATCAAGATTTTCTCATTAAAAAAATTCATTTTTCCTTTCTAATTTCATTTTTCTATCTTTTTTCCTTCAATTTTCCTTATTTTCATCTCTCTCTCTTCCTGTTTCTCTGAATTACCATAATTACCTCAAAAAATTGTGCAATAGTTTTACCCACTATTTTTTGTTCTCCAGCCCTTTCCCGAATCGATCATTTCTCTCTTTGTTGTCATTTTAAAAACATTTTGTCTTTTCTTTTTTGCTCTGAATTGTCTCCTCCAATTATTCAAATCTTGTGTCCCTAGAAAGATGAAAAAAAAAGTTATAAAAATAAATGGTTAATTGATTGATAGCACCTGCAACAAACAATTCAATGGGATATACAAGGCACTAAAAAAACTGGGTAGTTGGGGAATCTTTGTGAAAGAAAAACGGGGATGCAAAAAACATGAGATGTTACAGCAAAATTAAATGTCATGAAATTTCAAAATACCAGTAGAACACGTGAAAATGAGCTGCGTCAACGGCCCACCGGTAACAGAAAGAGAAAGATTCGAATGCAATAATTTCTAGATGCCATATCCTTGGTACTACTAGT
KI verification primers: SVH-5-CKIF: 5'-GAAGAGCAGAGTAAAAGTCTTGCACT-3' SVH-5-CKIR: 5'-AGTTTGAGGACATCTCCCTCGAC-3' Download
|
Strain name: SYS639 KI background: JIM113 Strain genotype: ujIs113 (pie-1p::mCherry::H2B; nhr-2p::mCherry::his-24::let858 3'UTR; unc-119(+)) II; svh-5 (devKi196 [svh-5::mNeonGreen]) X
|
Expression pattern: Expressed after bean stage
|
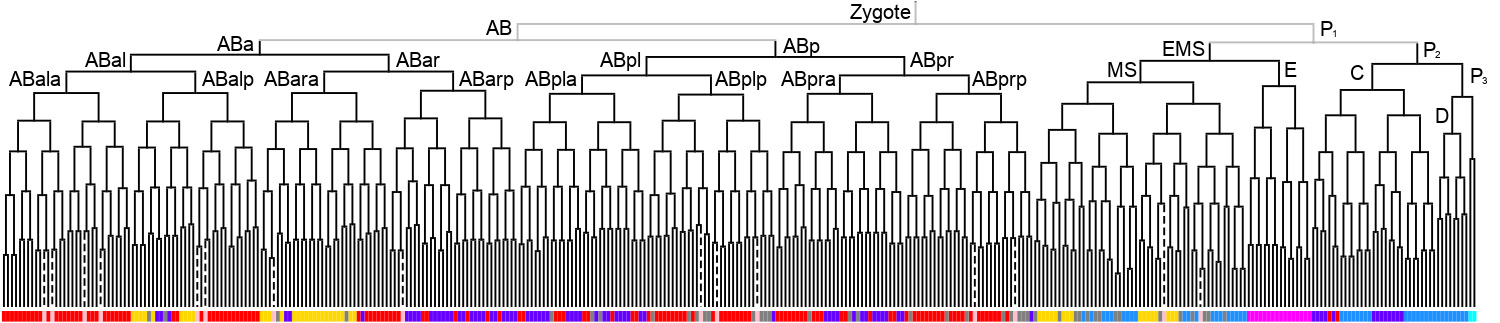
Lineage expression pattern
350 stage

|
600 stage
 |
3D expression pattern  4-cells 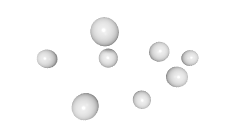 8-cells 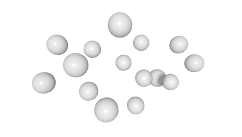 15-cells  28-cells  ~50-cells  ~100-cells  ~200-cells  ~350-cells  ~500-cells  ~600-cells  600-cells  bean-stage |
Inferred TF relationships
Based on expression similarity (strong, medium, weak): NA
Based on ChIP-seq (directly from ModERN database, targets from L1 stage ChIP-seq data are underlined)
Targeting: no-data
Targeted by: NA |
|
 Based on reannotated ChIP-seq (peaks are assigned to a target if it is located within 2 kb relative to the transcription start site; targets from L1 stage ChIP-seq data are underlined) Based on reannotated ChIP-seq (peaks are assigned to a target if it is located within 2 kb relative to the transcription start site; targets from L1 stage ChIP-seq data are underlined)
Targeting: no-data
Targeted by: BCL-11, CEH-14, CEH-18, CEH-31, CEH-34, ELT-2, HAM-2, HMG-4, M03D4.4, MLS-2, NFYA-1, ODD-2, PHA-4, REF-2, TBX-2, UNC-42
|














