WormBase ID: WBGene00004024 WormBase description: Is an ortholog of human HOXA10 (homeobox A10). Is predicted to have DNA binding activity. Is involved in positive regulation of nematode male tail tip morphogenesis. Is expressed in several structures, including coelomocyte; head neurons; intestine; somatic nervous system; and tail neurons. Human ortholog(s) of this gene are implicated in several diseases, including brachydactyly-syndactyly syndrome; diabetes mellitus (multiple); and dysostosis (multiple). |
Reporter verification: Verified by PCR and sequencing |
Strain name: SYS1013 Cross: RW12213 x JIM113 Strain genotype: ujIs113 (pie-1p::mCherry::H2B; nhr-2p::mCherry::his-24::let858 3'UTR; unc-119(+)) II; php-3(st12213[php-3::TY1::EGFP::3xFLAG]) III |
Expression pattern: Lineage-specific, Posterior-biased |
Lineage expression pattern 350 stage  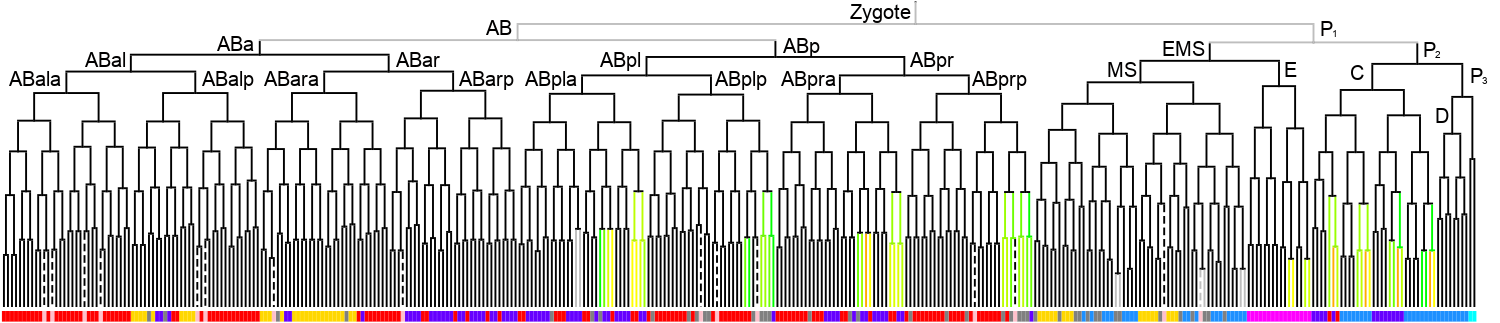 |
600 stage  |
3D expression pattern  4-cells 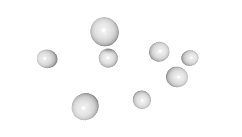 8-cells 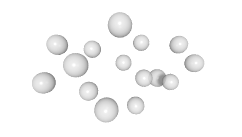 15-cells  28-cells  ~50-cells  ~100-cells  ~200-cells  ~350-cells  ~500-cells  ~600-cells 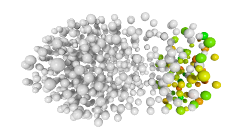 600-cells 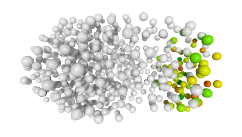 bean-stage |
Inferred TF relationships Based on expression similarity (strong, medium, weak): NOB-1, FOS-1, COG-1, VAB-15, VAB-7, EGL-5, UNC-3, TLP-1, PAL-1, NHR-25, LIN-26, NHR-23, NHR-67, NHR-102, HAM-2, HLH-14, CEH-20, HBL-1, LIN-1, ELT-1, UNC-120, Y75B8A.6, ETS-7 Based on ChIP-seq (directly from ModERN database, targets from L1 stage ChIP-seq data are underlined) Targeting: no-data Targeted by: NA |
|
Targeting: no-data Targeted by: NA |